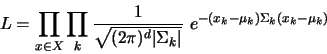 |
(2) |
The Mixture of Gaussians model represents a dataset by a set of mean and covariance matrices. Each class is centered at a mean and has a Gaussian which extends as described by it's matrix. Each class also has a weight associated with it which is simply its total fraction of points divided by the total number of points in the dataset.
The formula for computing the fitness of a dataset given a model is defined by
where ![]() is the mean of cluster
is the mean of cluster ![]() ,
, ![]() is the covariance matrix of cluster
is the covariance matrix of cluster ![]() ,
, ![]() is the dimensionality of the data, and
is the dimensionality of the data, and ![]() is the set of test datapoints.
is the set of test datapoints.
Typically, we compute the log-likelihood instead since the exponentiation in equation 2 will often exceed machine precision and the cumulative products will drive the answer to zero rather quickly. The formula for the likelihood then becomes
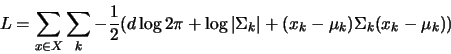 |
(3) |
Typical values for fitness will be on the order of
![]() . Now let's try an example where we divide the dataset in half putting even rows in one subset, odd rows in another. We will then cluster one of the subsets via the DiagEM wrapper (since it produces a Mixture of Gaussians model) and score the model based of the other subset.
. Now let's try an example where we divide the dataset in half putting even rows in one subset, odd rows in another. We will then cluster one of the subsets via the DiagEM wrapper (since it produces a Mixture of Gaussians model) and score the model based of the other subset.
>>>
>>> ds = Dataset('synth_t_15c3_p_0750_d_03_v_0d3.txt')
>>> evens = ds.subsetRows(range(0,750,2))
>>> odds = ds.subsetRows(range(1,750,2))
>>> parameters = {}
>>> parameters['k'] = 15
>>> parameters['num_iterations'] = 1000
>>> parameters['distance_metric'] = 'euclidean'
>>> parameters['init_method'] = 'church_means'
>>> diagem = DiagEM(evens, parameters)
>>> diagem.validate()
1
>>> diagem.run()
1
>>> model = diagem.getModel()
>>> model
MoG_Model(k=15, ...)
>>> model.evaluateFitness(odds.getData())
-963.03029940902013
The actual fitness number returned is meaningless on its own as it is a dimensionless number. Fitness scores are useful when used as relative comparisons. It is important to remember that since there is no intrinsic scale associated with a fitness score (it is model-dependent), you cannot say that a fitness of -500 is twice as good as a fitness of -1000. All you know is that it is a better score.