What next?
Diane Trout
2012-01-17
What next?
Table of Contents
- 1 Dangerous addresses
- 2 logins by status
- 3 "bad" by CC
- 4 "good" by CC
- 5 The Internet
- 6 File server from CACR
- 7 UID renumbering
- 8 Fileserver Status
- 9 Meta-data
- 10 Ontologies and you.
- 11 UCSC Uses meta-data for filtering
- 12 UCSC detailed meta-data
- 13 Meta-data we capture
- 14 Meta-data I fill in later
- 15 LHCN
- 16 Current processing.
- 17 Meta-data limits
- 18 UCSC Updates
- 19 Human ChIP-Seq
- 20 Single Cell RNA-Seq
- 21 Mouse Freeze
- 22 HTSW/Tracking data
- 23 High priority HTSW bugs
- 24 Future directions
- 25 Who knows if
- 26 More QA
- 27 Shared QA
- 28 Experiment QA
- 29 QA UI
- 30 QA UI
- 31 Many ways to accomplish that.
- 32 Wiki
- 33 Wiki for QC
- 34 Simpler path
- 35 PingBack
- 36 Requirements, I think
- 37 Jumpgate addition
- 38 Note taking
- 39 Wiki as note taking
- 40 Code + version control
- 41 Project Site?
- 42 GANTT Charts
- 43 Galaxy?
- 44 MS Office
- 45 SharePoint
- 46 iPython Notebook?
- 47 Org-mode
- 48 Blogs
- 49 TOO MANY CHOICES
- 50 What have you used?
- 51 How are you actually taking notes?
1 Dangerous addresses
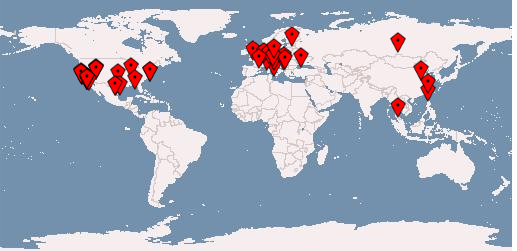
2 logins by status
- 54,564 "successful"
- 339,216 "errors"
- (not all are hostile, and not all are separate events, my log analyzing skills need improvement)
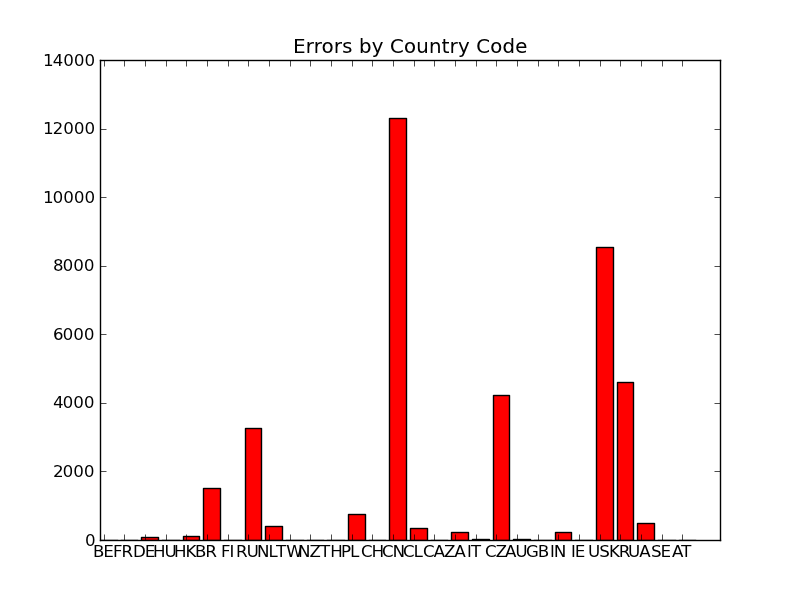
3 "bad" by CC
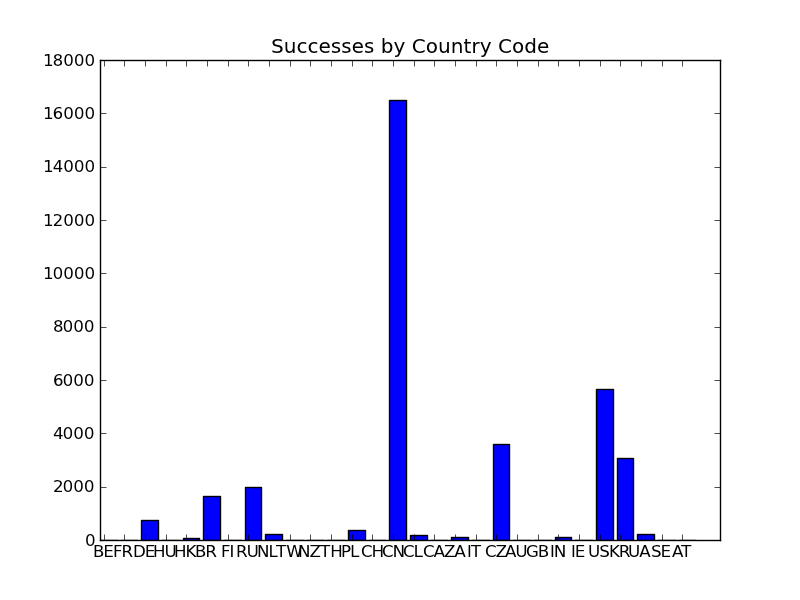
4 "good" by CC
5 The Internet
- Its like Swimming in a disease infested swamp.
- Make sure you don't have open wounds.
- don't run unnecessary services
- keep software up to date – especially things that use the network
- Analyzed data was from Dec 11 06:28:51 - Jan 13 11:50:22
- Only ssh – and we have throttling turned on.
6 File server from CACR
- Took way too long to order.
- The initial expectations wouldn't work
- Needed several meetings to figure out how to share
7 UID renumbering
- Side effect, we will need to renumber our user IDs to be compatible with CACR
8 Fileserver Status
- Henry is using the new server to try move things off of bad disks so we can reformat or replace with larger disks.
- (Can we buy more disks for the genome browser? Currently 1TB disks, would like to replace ~15 disks with 2TB disks.
Though right now still isn't the best time to buy disks. prices are still high due to flooding.
"With operations disrupted at more than a dozen hard disk drive (HDD) factories due to flooding in Thailand, PC manufacturers should prepare for significant supply shortages, market research firm IDC said Thursday." - computer world Nov 10, 2011
9 Meta-data
- It's actually important
10 Ontologies and you.
- UCSC is trying to get us to describe our samples using a common vocabulary.
11 UCSC Uses meta-data for filtering
12 UCSC detailed meta-data
GM12878 single read RNA-seq Minus Signal Rep 1 from ENCODE/Caltech
shortLabel: GM78 1x75D - 1
Principal Investigator on grant: Myers
Lab producing data: Wold - CalTech
Experiment (Assay) type: RNA-seq
View - Peaks or Signals: Raw signal -
Experiment ID: 125
Cell, tissue or DNA sample: GM12878
Cellular compartment: Whole cell
RNA Extract: Long PolyA+ RNA
Paired/Single reads lengths: 1x75D
Replicate number: 1
Version of data if resubmitted: V3 - A better mapping technique.
Assembly originally mapped to: hg18
ENCODE Data Freeze: post ENCODE Jan 2011 Freeze
UCSC Accession: wgEncodeEH000125
Date submitted to UCSC: 2010-01-04
Date resubmitted to UCSC: 2011-02-14
Date restrictions end: 2010-10-04
Submission ID: 3514
Lab provided identifier: 11011
Mapping algorithm: TH1014
tableName: wgEncodeCaltechRnaSeqGm12878R1x75dMinusSignalRep1V3
fileName: wgEncodeCaltechRnaSeqGm12878R1x75dMinusSignalRep1V3.bigWig
13 Meta-data we capture
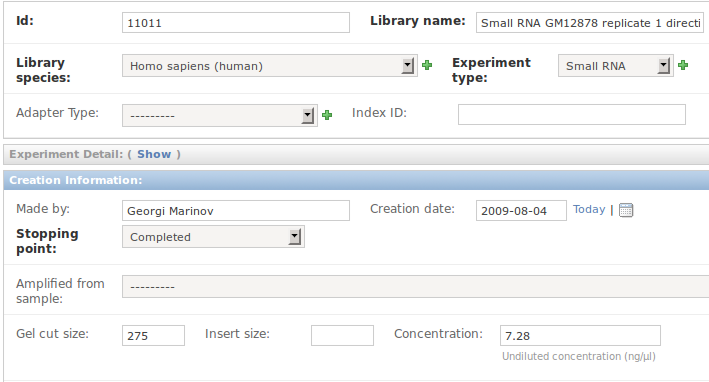
14 Meta-data I fill in later
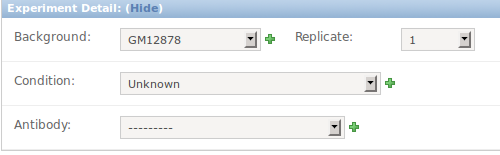
15 LHCN
- We call it LHCN-cycling vs LHCN-differentiated
- UCSC/NIH decided there is only the LHCN cell line
- It has two treatments None, and 7 day differentiated
- If you have admin priviledges on jumpgate, use LHCN
16 Current processing.
- Georgi preps an analysis
- hands files to me
- I figure out what meta-data UCSC wants, and try to shove into databases
- Generate meta-data files
- upload
- discuss what went wrong.
17 Meta-data limits
- Additional Meta-data is ENCODE specific
- Labs in ENCODE have enough trouble agreeing on terms.
- Igor pointed out worm and chemical assay groups would have wildly different needs.
18 UCSC Updates
- Results of recent meeting with Cricket/Venkat
19 Human ChIP-Seq
20 Single Cell RNA-Seq
- According to Cricket, Peter Goode said it wasn't core
- so UCSC wasn't planning on accepting it
- Perhaps we would be interested in uploading to GEOs UCSC page?
- (Not much visualization, just a place to download data)
- If this is not acceptable someone needs to get Peter Goode to tell Cricket to accept the data.
21 Mouse Freeze
March 31
- What we were supposed to be submitting
- Who is the responsible person for generating samples
- Are they on these lists?
- Encode
- Mouse Encode
22 HTSW/Tracking data
- Whats next for tracking our data & processing
23 High priority HTSW bugs
- extract results for HiSeq runs.
- (well first define what run information to save from HiSeq)
- display HiSeq summary reports.
24 Future directions
The biggest problem(1) our lab faces is sharing knowledge about our experiments
(1) (that I have a chance of doing anything about)
25 Who knows if
- Library 11011 is it good or not?
- 10155?
26 More QA
The jumpgate database is tracking "samples" handled by the gene expression center. Some QA features apply to all experiment types, but some are specific to the lab & experiment.
There's also disagreement about the validity of a QA method.
27 Shared QA
- Reports generated by pipeline
- e.g. intensity plots
- read counts
- report average quality scores
- many other things I don't know about?
28 Experiment QA
- Uniformity measurement
- 5'/3' read distribution
- Reads aligned to introns
29 QA UI
- Adding to jumpgate would be slow & cluttered
- bolting on random features leads to clunky software
- see MS Word, iTunes
- it might work if jumpgate was just for us, but other labs will have their own needs, and our analysis may or may not apply.
- Not only does it makes me a bottle-neck, I'm not qualified to pick among the QC methods
30 QA UI
- What we want is a central place to find information
about an experiment.
- not a just, is a sample good
- but, why did we make it?
- is it related to another sample?
- what analysis has been done on it.
31 Many ways to accomplish that.
- Ye olden days had meetings with shouting over mead and pointing
at each other with haunches of meat…
- and secretaries who took notes and mailed them to people
- Has issue in the modern distributed workforce
- Also organizations are prone to forgetting.
32 Wiki
- https://woldlab.caltech.edu/wiki/RecentChanges
- https://jumpgate.caltech.edu/wiki/RecentChanges
- You all can edit the first site, and probably the second site.
- Its mostly been me and henry who've edited.
- The encode wiki gets more use.
- http://encodewiki.ucsc.edu/EncodeDCC/index.php/Special:RecentChanges
33 Wiki for QC
Tried to propose:
http://jumpgate.caltech.edu/wiki/Library/10155
- Would need a few custom macros to "transclude" jumpgate data
- I'm not good at getting buy in.
34 Simpler path
A directory
- write reports to directory
- Like 70JBCAAXX
35 PingBack
- Another idea is to make the library page aware of people linking to it
- e.g. 10155-example.html
36 Requirements, I think
- An accessible, usable place to write about an experiment
- A Willingness to write things down.
- It needs to be searchable
- It needs to support showing text, plots, tables, images.
- Physical objects?
- It should allow sharing and reusing code.
37 Jumpgate addition
- Jumpgate needs improvements in letting downstream users know there's new data.
- Subscribe to a library?
- Get an email/im/RSS entry every time new data is added?
38 Note taking
After enough abstraction the QC problem turns into
- How do we take notes and share them.
39 Wiki as note taking
see earlier Wiki
40 Code + version control
- Tracks process well
- I've seen projects that you could unarchive, type "make paper" and re-run the analysis and get the same paper out the other end.
- Great for tracking processes, less good for why? (Unless you aim for "$ make paper")
- Titus seems to be using it for his lab
- (Also what about relationships between datasets?)
41 Project Site?
Like: https://mus.cacr.caltech.edu/htsworkflow/
- Its a wiki, but adds ticket tracking
- can be linked to show source code.
- And maybe a gantt chart
42 GANTT Charts
(aside)

- Imagine deadlines in a gantt chart.
- Imagine showing estimated duration for sequencing and analysis.
43 Galaxy?
- web based tool for managing experiments
- http://galaxyproject.org
- http://main.g2.bx.psu.edu/u/nickpolato/h/apalmataassembly
- Sysadmins were resistant because we don't think we have a workable database server.
44 MS Office
- many people find easy to use
- information gets locked up on laptops
- tracking versions is hard. (3-way merge is by hand)
- or maybe not? google turned up subversion for word files?
45 SharePoint
- server to host MS Office content
- "content management"
- "document management"
- My knowledge is limited
- other, similar, tools might even work
46 iPython Notebook?
- It's python mathematica in a web page!
- Titus & iPython developers are thinking about developing
workflows in ipython notebook.
- I think this'd be develop a notebook to analyze a dataset
- The start would have a header defining where the dataset is located
- Copy notebook, change input?
47 Org-mode
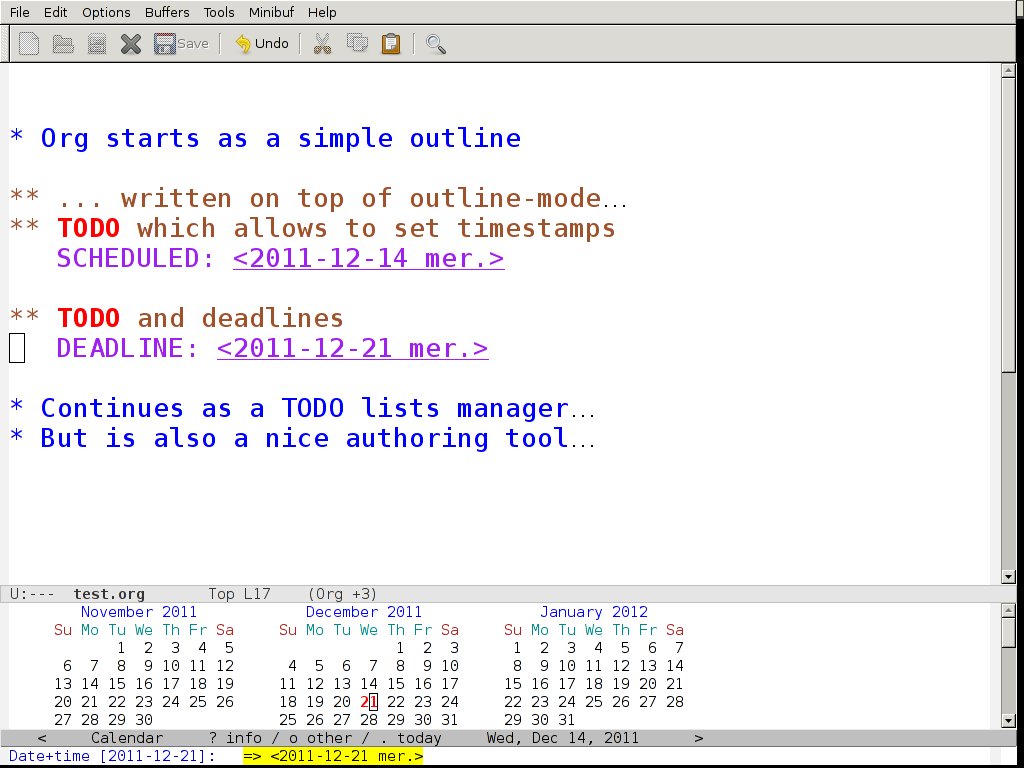
- http://orgmode.org
- Its everything in an outline in Emacs.
48 Blogs
- Instead of organized around a topic like a wiki, blogs are date
- http://ivory.idyll.org/blog/
49 TOO MANY CHOICES
- I'm bad at picking
- between lots of functionally similar choices.
- things that other people will willingly use.
50 What have you used?
- Blog
- Wiki
- twitter/facebook/google+
- RSS Reader
- Mailing list.
51 How are you actually taking notes?
- You are taking notes aren't you?